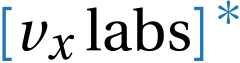Contact QRCode generator with marimo and WASM
Introduction
Just the other weekend, I had to exchange contact details using quite primitive means with a fantastic new friend after a deeply enjoyable hike up Table Mountain via the India Venster route.
We could not get the flashy new iPhone-bump method to work. Although the animation triggered with each bump, no contact details were exchanged, a failure which caused the Android user at the table much mirth.
That incident drove me to retrieve some old Python code I had written to try out Segno, a QRCode generator library for Python, and to test out the recently announced WASM capability of the new Marimo reactive Python notebook.