Speed up Obsidian Quartz page loads
I recently setup Quartz to publish a subset of my “howto” notes to the TiL section of my website.
Today I had some time to dive (delve…. haha) into why the page loads did not feel as fast as I am used to from my other static sites.
PageSpeed Insights
PageSpeed Insights reported that mobile page speed performance was 34 and desktop better but not great at 81:
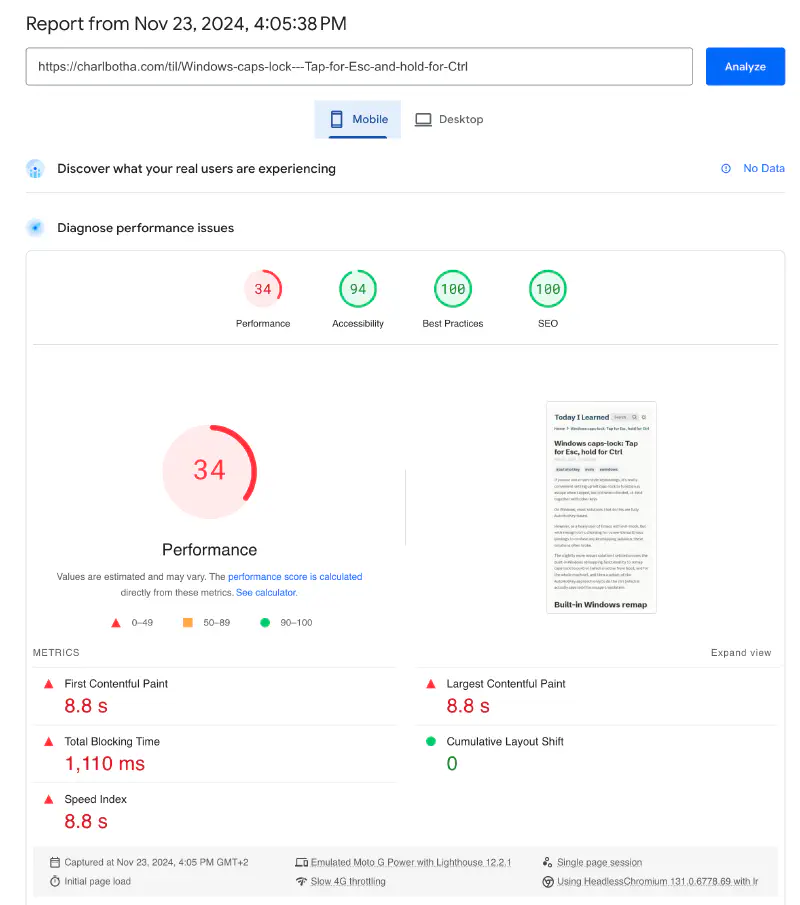
Let’s take a look inside the bundle
First thing I checked was the browser’s dev tools. Here we can see that the HTML page itself is far too large at 850kB and that postscript.js not far behind at 224kB.