Performance comparison of six different LTTB (visual downsampling for timeseries data) algorithm implementations for Python
Contents
LTTB, or Largest-Triangle-Three-Buckets, is a fantastic little algorithm that you can use for the visual downsampling of timeseries data.
Let’s say your user is viewing a line chart of some timeseries data. The time-period they have selected contains 50000 points, but their display is only 4K so they have a maximum of 3840 pixels available horizontally. With LTTB, we can automatically select the 3840 or fewer points from those 50000 points that will produce a line graph which is visually very similar to what they would see if they were to try and render all 50000 points.
This is really great, because now they only have to download and render those 3840 points, contributing to much faster response times.
Below you can see some more modest examples generated from a demo timeseries of 5000 points.
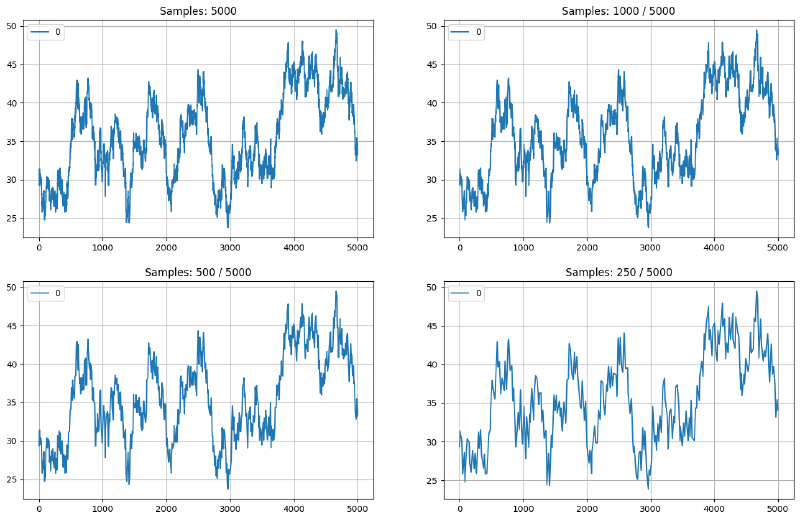
Figure 1: Example of LTTB algorithm: 5000 input samples downsampled to 1000, 500 and 250. Line plots maintain visual similarity to original.
Do also try out the interactive demo at https://www.base.is/flot/
Besides the usefulness of the algorithm itself, what I really love is that it had a humble start as the topic of Sveinn Steinarsson’s 2013 CS Master’s thesis work but has now been translated into a long list of programming languages and it really looks like it has had significant impact on the computing world!
Anyways, the topic of this post is the following: If you wish to serve LTTB-downsampled data from your Python service, what are your options, and how fast are they? In other words, we are going to do some backyard benchmarking!
Besides the comparison, this work offers two additional contributions:
- The first publically available Pythran (Python to C++ transpiler) tweaking of javiljoen’s lttb-numpy implementation, thanks to a PR by the indomitable @stefanv.
- The first publically available Cython implementation of the LTTB algorithm which performs to within 10% of the fastest C implementation.
Show me the code
In the lttb-bench github repo you’ll find hopefully reproducible demonstrations of the following implementations and variations of those implementations:
- lttbc: Implementation in C using the numpy API
- lttb-numpy: Pure Python implementation that uses numpy
- lttb-pythran: Slightly tweaked version of the above to enable Pythran conversion
- ltttb-numba: Slightly tweaked version to enable numba conversion
- pylttb: Alternative pure Python and numpy implementation
- lttb-cython: Cython implementation, which calls this repo its home
In all cases, start by going through the Jupyter notebook. If you’re pressed for time, you can jump to the last cell which contains all code to prepare and run all six of the implementations.
Lies, damned lies and benchmarks
See the table below for the results of benchmarking the six variants using the %timeit notebook magic with mostly default settings, i.e. automatically looping the implementation enough times for accurate measurements, and calculating the mean and standard deviation over 7 runs of N loops each. All times are in µs (micro-seconds) per loop.
| Platform | C | cython | pythran | numba | lttb-numpy | pylttb |
|---|---|---|---|---|---|---|
| Ryzen 9 5900x WSL Ubuntu 22.04 | 10.6 | 11.8 | 31.6 | 118 | 3860 | 3700 |
| Ryzen 9 5900x Windows 11 VS 2022 | 10.5 | 11.8 | 23.3 | 144 | 3370 | 3830 |
| MacBook M1 Pro / Max MacOS 14.4.1 | 19.9 | 21.3 | 16.7 | 178 | 2250 | 2400 |
Some observations:
- The algorithm is quite sequential, so it can’t be effectively vectorized and the Python / numpy implementations are much slower than the compiled versions.
- My Cython implementation is within 10% of the C implementation, which is nice, because writing Cython is often easier than C.
- Surprisingly, the Pythran (Python to C++ conversion) is faster than the hand-coded C on Mac.
- numba gives us quite a lot of performance for very little effort and complexity. What I mean by that is that for the C, Cython and Pythran implementations you need a compiler toolchain on the machine that’s building your packages. numba depends on llvmlite to perform jit compilation of your Python code, no compiler toolchain required and also no programming in a different language (C, Cython) required.
Conclusion
I had fun comparing these LTTB implementations, and even more fun making the Cython one work.
If I had to use lttb in production (again…) I would stick with the more flexible and easy to build but slower versions.
For example, in our production journey we realized only later that we would also need the actual indices of the samples that were selected, and so I had to hack the lttbc code to return that information. You can find that work in my cpb-return-offsets fork of the original lttbc. This will probably not make it back upstream, and that’s part of the problem.
Although Cython and even more so Pythran are huge fun and more flexible than using the numpy C API, do make sure that your build processes are fine maintaining these extra steps to get the high-performance binaries built for your deployment platforms.
Numba looks like a great solution that makes a different tradeoff: You get maximum flexibility and no compilation steps required during deployment, but you can get way better performance than pure Python although you’re coding in more-or-less pure Python.