Run code on remote ipython kernels with Emacs and orgmode.
Contents
As is briefly documented on the ob-ipython github, one can run code on remote ipython kernels.
In this post, I give a little more detail, and show that this also works wonderfully for remote generation but local embedding of graphics in Emacs Org mode.
As I hinted previously, the jupyter notebook is a great interface for computational coding, but Emacs and Org mode offer far more flexible editing and are more robust as a documentation format.
On to the show. (This whole blog post is a single Org mode file with org-babel source code blocks, the last of which is live.)
Start by starting the ipython kernel on the remote machine:
me@server$ jupyter --runtime-dir >>> /run/user/1000/jupyterme@server$ ipython kernel >>> To connect another client to this kernel, use: >>> –existing kernel-11925.json
We have to copy that json connection file to the client machine, and then connect to it with the jupyter console:
me@client$ jupyter --runtime-dir >>> /Users/cpbotha/Library/Jupyter/runtimeme@client$ cd /Users/cpbotha/Library/Jupyter/runtime me@client$ scp me@server:/run/user/1000/jupyter/kernel-11925.json . me@client$ jupyter console –existing kernel-12818.json –ssh meepz97 >>> [ZMQTerminalIPythonApp] To connect another client via this tunnel, use: >>> [ZMQTerminalIPythonApp] –existing kernel-12818-ssh.json
Note that we copy the json file into our local jupyter runtime directory, which will create the ssh connection file there, and enable us to reference it by name.json only (vs its full path) in any ob-ipython source code blocks.
Now you can open ob-ipython org-babel source blocks which will connect to the remote kernel. They start like this:
#+BEGIN_SRC ipython :session kernel-12818-ssh.json :exports both :results raw drawer
Let’s try it out:
%matplotlib inline # changed to png only for the blog post %config InlineBackend.figure_format = 'png'from mpl_toolkits.mplot3d import axes3d import matplotlib.pyplot as plt from matplotlib import cm
fig = plt.figure() ax = fig.add_subplot(111, projection=‘3d’) X, Y, Z = axes3d.get_test_data(0.05) cset = ax.contour(X, Y, Z, cmap=cm.coolwarm) ax.clabel(cset, fontsize=9, inline=1)
plt.show()
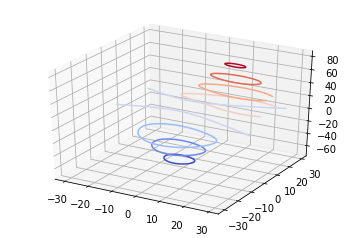
Pretty amazing! The code is executed on the remote machine, and the resultant plot is piped back and displayed embedded in orgmode as SVG!